Motivation: Much of the difficulty in rendering consistent evaluation of pathological tissue specimens arises from the qualitative impressions of observers. Hence, a quantitative phenotype to automatically characterize the type and stage of cancer based upon an objective measure of its morphology and mechanical properties could provide insight regarding the changes that occur in the tissue environment during the course of disease onset and progression. We hypothesize that contact-mode Atomic Force Microscopy (AFM) could be used to quantify the mechanical properties of breast tissue undergoing morphological transformation due to increasing malignancy. We use breast tissue cores fixed on microscope slides using Tissue Microarray Technology (TMA) during AFM based experiments. The salient features of this project comprise: (a) development of an AFM system with high-throughput raster indentation capability and (b) development of computational tools to facilitate accurate mechanical property estimation from raw AFM data.
Project Highlights:
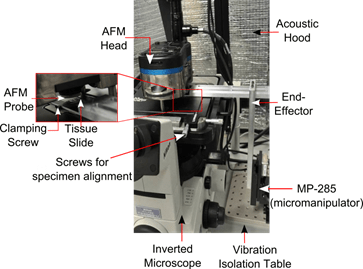
Automated vision-guided navigation of TMA slide underneath AFM tip
While commercial AFM’s are capable of measuring forces with nN resolution, they are limited by their scanner’s travel range. Conventional AFM scanners have around 100 μm X-Y range for closed-loop positioning, while TMA cores are typically sectioned at 600 μm diameter, primarily to ensure that malignancies of interest are sufficiently represented on a tissue specimen. This necessitates repetitive manual alignment of the AFM probe and tissue specimens, which severely restricts throughput. Precise probe-tissue alignment using manual methods also increases AFM operator fatigue and these factors call for increased automation in carrying out AFM indentations on tissue specimens.
To this end, we have developed an image-guided positioning system for precise alignment of the AFM probe tip and tissue specimens as large as 600 μm in diameter. Online tracking during positioning was performed by implementing a template matching algorithm on the “Tracking ROI”, while the “Probing ROI” was raster scanned by the AFM probe. The positioning system is also capable of performing tissue registration across multiple magnifications, and this system significantly improves throughout and reduces operator fatigue.
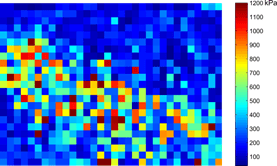
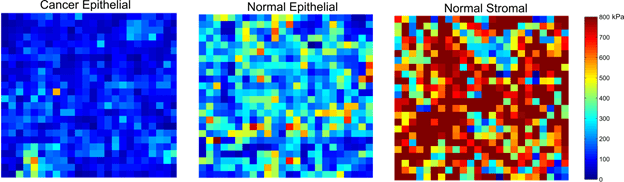
Our initial results indicate that cancerous epithelial tissue display reduced elastic modulus compared to normal epithelial tissue. We also observed that stromal regions are stiffer than epithelial regions, potentially due to the domination of fibrous tissue in these regions.
Uncertainty estimation in AFM measurements
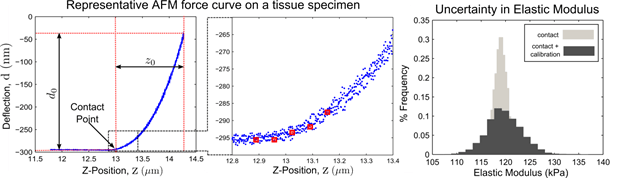
In contact mode operation of the AFM, the force (F) is obtained as a product of the AFM probe deflection (d) and its spring constant (k), which is typically calibrated at the start of an AFM experiment. The force is then recorded as a function of the probe’s vertical position (z). This process results in a force-displacement (F-z) (typically called an AFM force curve). To extract the elastic modulus from the AFM force curve, the force is required as a function of the sample indentation. This is achieved by first identifying the point on the force curve where the probe touches the sample, called the contact point. The sample indentation is then obtained by subtracting the post contact deflection (d0) from the post-contact vertical travel (z0). The indentation force (F0 = kd0) thus obtained as a function of the indentation (z0 – d0) is subsequently fitted to a contact model to estimate the specimen’s elastic properties.
Unfortuntely, the AFM experimental parameters involved in the mechanical characterization process are rarely obtained error-free. The first source of error stems from uncertainty in the location of the contact point. This is fundamentally due to the smooth transition from the non-contact to the contact regime in AFM force curves on compliant specimens, which leads to uncertainty in the parameters (d0) and (z0). Furthermore, the spring constant calibration typically shows variations in the order of 5-17%, leading to further uncertainty in (F0).
Our research focuses on development of a robust mathematical model that accurately captures the uncertainty due to the aforementioned factors. To this end, we have developed an Error-In-Variables (EIV) based Bayesian Changepoint algorithm to investigate the impact of these uncertainties in the mechanical characterization process. We have validated our approach to simulated data, and the EIV approach can successfully capture variability in the probe calibration measuments.
Personnel: Rajarshi Roy
Sponsor: NIH R01 Grant.
Publications:
- Rajarshi Roy, Wenjin Chen, Lauri Goodell, Jun Hu, David Foran, Jaydev P. Desai, “Microarray-facilitated Mechanical Characterization of Breast Tissue Pathology Samples using contact mode Atomic Force Microscopy (AFM),” in Third IEEE/RAS-EMBS International Conference on Biomedical Robotics and Biomechatronics – Biorob 2010, Tokyo, Japan, September 26-29, 2010
- Rajarshi Roy, Wenjin Chen, Lei Cong, Lauri Goodell, David Foran, Jaydev P. Desai, “Towards Automated Mechanical Characterization of Biological Samples Using Atomic Force Microscopy (AFM),” in 4th Annual Dynamic Systems and Control Conference, Oct 31 – Nov 2, 2011, Arlington, VA, USA
- Rajarshi Roy, Lei Li, Carol Keefer, Jaydev P. Desai, “An Error-In-Variables (EIV) based Bayesian Probabilistic Approach to Estimating Cell Mechanical Properties Using Atomic Force Microscopy,” in Third IEEE/RAS-EMBS International Conference on Biomedical Robotics and Biomechatronics – BioRob 2012, Roma, Italy. June 24-27, 2012
- Rajarshi Roy, Wenjin Chen, Lei Cong, Lauri Goodell, David J. Foran, and Jaydev P. Desai, “A
Semi-Automated Positioning System for contact-mode Atomic Force Microscopy (AFM),” IEEE Transactions on Automation Science and Engineering, vol. 10, no. 2 pp 462-465, 2013. - Rajarshi Roy, Wenjin Chen, Lei Cong, Lauri Goodell, David J. Foran, and Jaydev P. Desai, “Probabilistic Estimation of Mechanical Properties of Biomaterials Using Atomic Force Microscopy”, IEEE Transactions on Biomedical Engineering, (In Press), 2013.